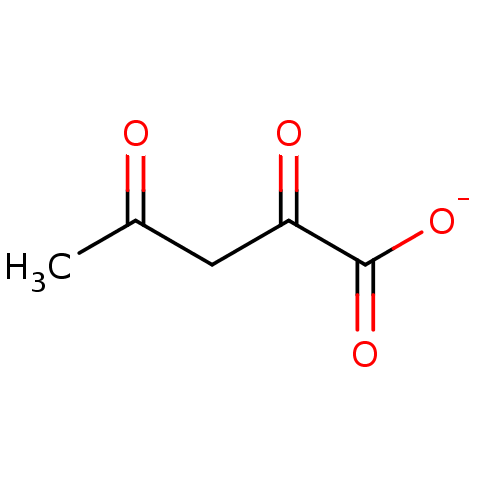
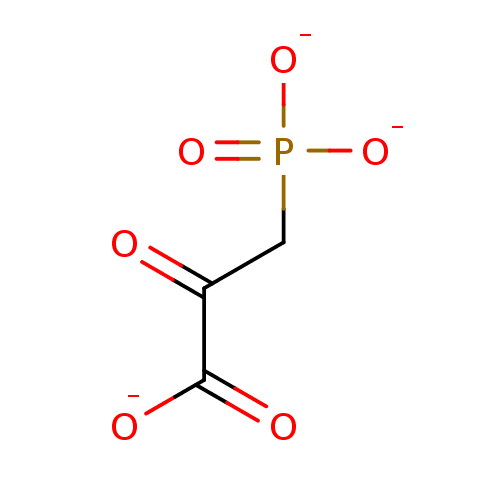
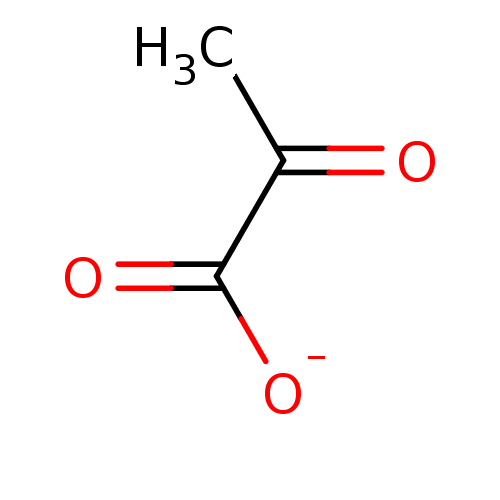
TargetOxaloacetate decarboxylase(Pseudomonas aeruginosa (strain ATCC 15692 / PAO1 /...)
University of Maryland Biotechnology Institute
University of Maryland Biotechnology Institute
Affinity DataKi: 4.30E+4nMAssay Description:Competitive inhibition constant determined for PA4872 catalyzed oxaloacetate decarboxylation.More data for this Ligand-Target Pair
TargetOxaloacetate decarboxylase(Pseudomonas aeruginosa (strain ATCC 15692 / PAO1 /...)
University of Maryland Biotechnology Institute
University of Maryland Biotechnology Institute
Affinity DataKi: 4.50E+5nMAssay Description:Competitive inhibition constant determined for PA4872 catalyzed oxaloacetate decarboxylation.More data for this Ligand-Target Pair
TargetOxaloacetate decarboxylase(Pseudomonas aeruginosa (strain ATCC 15692 / PAO1 /...)
University of Maryland Biotechnology Institute
University of Maryland Biotechnology Institute
Affinity DataKi: 5.60E+5nMAssay Description:Competitive inhibition constant determined for PA4872 catalyzed oxaloacetate decarboxylation.More data for this Ligand-Target Pair
TargetOxaloacetate decarboxylase(Pseudomonas aeruginosa (strain ATCC 15692 / PAO1 /...)
University of Maryland Biotechnology Institute
University of Maryland Biotechnology Institute
Affinity DataKi: 1.09E+6nMAssay Description:Competitive inhibition constant determined for PA4872 catalyzed oxaloacetate decarboxylation.More data for this Ligand-Target Pair
TargetOxaloacetate decarboxylase(Pseudomonas aeruginosa (strain ATCC 15692 / PAO1 /...)
University of Maryland Biotechnology Institute
University of Maryland Biotechnology Institute
Affinity DataKi: 3.00E+6nMAssay Description:Competitive inhibition constant determined for PA4872 catalyzed oxaloacetate decarboxylation.More data for this Ligand-Target Pair
TargetOxaloacetate decarboxylase(Pseudomonas aeruginosa (strain ATCC 15692 / PAO1 /...)
University of Maryland Biotechnology Institute
University of Maryland Biotechnology Institute
Affinity DataKi: 3.20E+6nMAssay Description:Competitive inhibition constant determined for PA4872 catalyzed oxaloacetate decarboxylation.More data for this Ligand-Target Pair
TargetOxaloacetate decarboxylase(Pseudomonas aeruginosa (strain ATCC 15692 / PAO1 /...)
University of Maryland Biotechnology Institute
University of Maryland Biotechnology Institute
Affinity DataKi: 6.70E+6nMAssay Description:Competitive inhibition constant determined for PA4872 catalyzed oxaloacetate decarboxylation.More data for this Ligand-Target Pair
TargetOxaloacetate decarboxylase(Pseudomonas aeruginosa (strain ATCC 15692 / PAO1 /...)
University of Maryland Biotechnology Institute
University of Maryland Biotechnology Institute
Affinity DataKi: 7.20E+6nMAssay Description:Competitive inhibition constant determined for PA4872 catalyzed oxaloacetate decarboxylation.More data for this Ligand-Target Pair

 3D Structure (crystal)
3D Structure (crystal)